Medical SAM 2: Segment Medical Images as Video via Segment Anything Model 2
Abstract
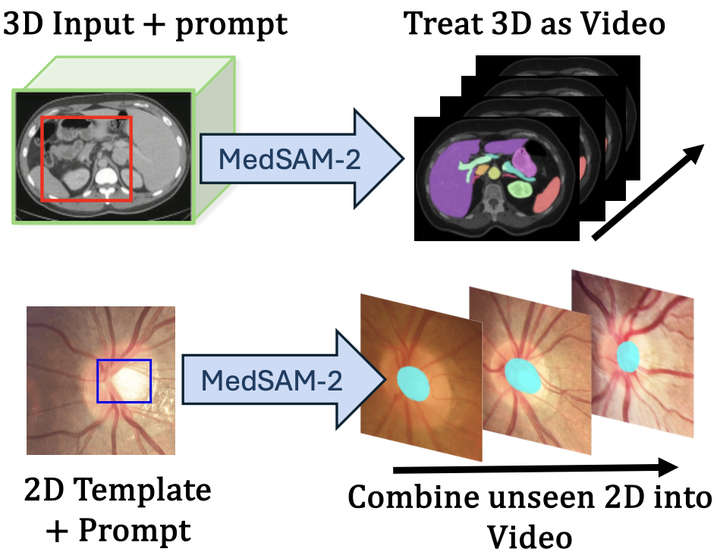
Medical image segmentation plays a pivotal role in clinical diagnostics and treatment planning, yet existing models often face challenges in generalization and in handling both 2D and 3D data uniformly. In this paper, we introduce Medical SAM 2 (MedSAM-2), a generalized auto-tracking model for universal 2D and 3D medical image segmentation. The core concept is to leverage the Segment Anything Model 2 (SAM2) pipeline to treat all 2D and 3D medical segmentation tasks as a video object tracking problem. To put it into practice, we propose a novel self-sorting memory bank mechanism that dynamically selects informative embeddings based on confidence and dissimilarity, regardless of temporal order. This mechanism not only significantly improves performance in 3D medical image segmentation but also unlocks a One-Prompt Segmentation capability for 2D images, allowing segmentation across multiple images from a single prompt without temporal relationships. We evaluated MedSAM-2 on five 2D tasks and nine 3D tasks, including white blood cells, optic cups, retinal vessels, mandibles, coronary arteries, kidney tumors, liver tumors, breast cancer, nasopharynx cancer, vestibular schwannoma, mediastinal lymph nodules, cerebral artery, inferior alveolar nerve, and abdominal organs, comparing it against state-of-the-art (SOTA) models in task-tailored, general and interactive segmentation settings. Our findings demonstrate that MedSAM-2 surpasses a wide range of existing models and updates new SOTA on several benchmarks.